
Help
About trRosettaRNA
The input to trRosettaRNA is the nucleotide sequence or a multiple sequence alignment (MSA) of the query RNA. Shown in Figure 1a, the trRosettaRNA works as follows.(1) When the sequence of query RNA or MSA is submitted, an attention-based nerual network (Figure 1b) is applied to predict the inter-nuclotide distance distribtuions.
(2) The predicted distance distribtuions are converted into smooth restraints, which are used to guide the Rosetta to build 3D structure models based direct energy mimization.
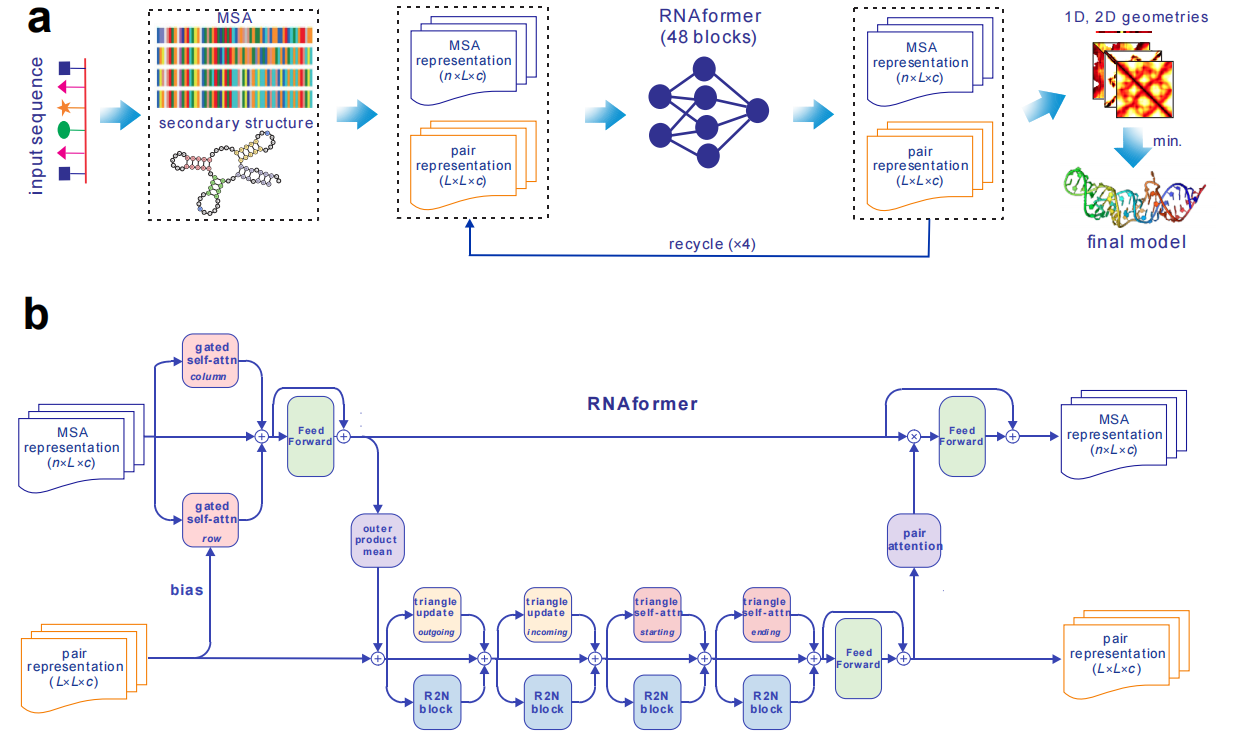
Figure 1. The flowchart and network architecture of the trRosettaRNA algorithm.
trRosettaRNA has been validated in blind tests, including CASP15 and RNA-Puzzles (Figure 2), which suggests that that the automated predictions by trRosettaRNA are competitive to the predictions by the top human groups on natural RNAs.

Figure 2. Blind test results in CASP15 and RNA-Puzzles.
The trRosettaRNA server runs BLASTN and Infernal on the RNAcentral database to generate MSA.
Based on the probability of the top predicted distance and the convergence of the top models, we estimate the RMSD of the predicted models. An RMSD lower than 4 Å usually indicates a model with correctly predicted topology.
Figure 3 shows the relationship between the estimated and the real RMSD for modeling on 50 RNAs used for benchmark tests. The Person's correlation coefficients is 0.56.

Figure 3. The relationship between the estimated RMSD and the real RMSD.
Job submission
Job submission guide:(1) Input a RNA sequence in FASTA format or a MSA in A3M/FASTA/A2M/STO format.
(2) Specify your input type.
(3) (Optional) Provide your custom secondary structure (If not provided, the secondary structure will be predicted by SPOT-RNA).
(4) (Optional) Provide your email address.
(5) (Optional) Assign your target name.
(6) Choose whether to keep your results private.
(7) Submit.
Notice: Due to computing resource limitation, we now allow no more than 20 running/pending jobs per user at the same time.

Figure 4. The "Submit" section in trRosettaRNA home page.
Output explanation
The trRosettaRNA modeling results are generally summarized in a webpage, the link of which is sent to the user upon job completion if the email address has been provided during submission(see an example of the trRosettaRNA output). A tallbar file containing the key modeling results can be downloaded from the top of this page.This section contains:
(1) Top-predicted model visualization.
(2) Model quality estimation.
(3) Modeling method description.
(4) Separate download links for models, MSA (multiple sequence alignment), inter-nucleotide distances.

Figure 5. The "Predicted Structure Models" section in trRosettaRNA result page.
This section visualizes predicted 2D information including:
(1) Contact map, which displays the predicted probability of nucleotide pairs being in contact, i.e., the minimum distance between all atoms of them is less than 8Å.
(2) Distance map, which displays the predicted real distance (3 - 40 Å) between nucleotide pairs.
(1) Contact map, which displays the predicted probability of nucleotide pairs being in contact, i.e., the minimum distance between all atoms of them is less than 8Å.
(2) Distance map, which displays the predicted real distance (3 - 40 Å) between nucleotide pairs.

Figure 6. The "Predicted 2D Information" section in trRosettaRNA result page.
This section visualizes the secondary structures including those:
(1) predicted by SPOT-RNA or input by user.
(2) extracted from model1.pdb.
(1) predicted by SPOT-RNA or input by user.
(2) extracted from model1.pdb.
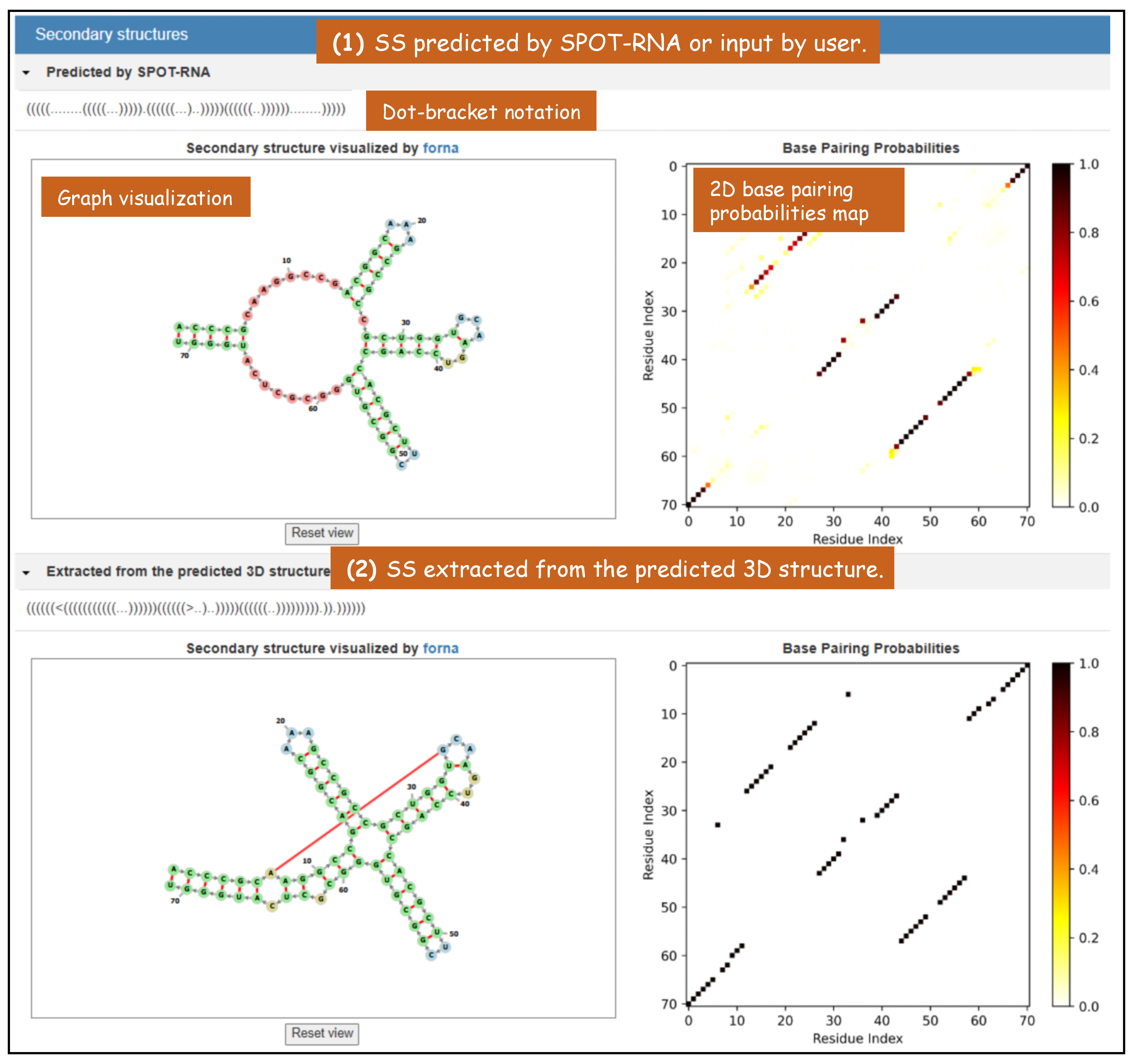
Figure 7. The "Secondary structures" section in trRosettaRNA result page.